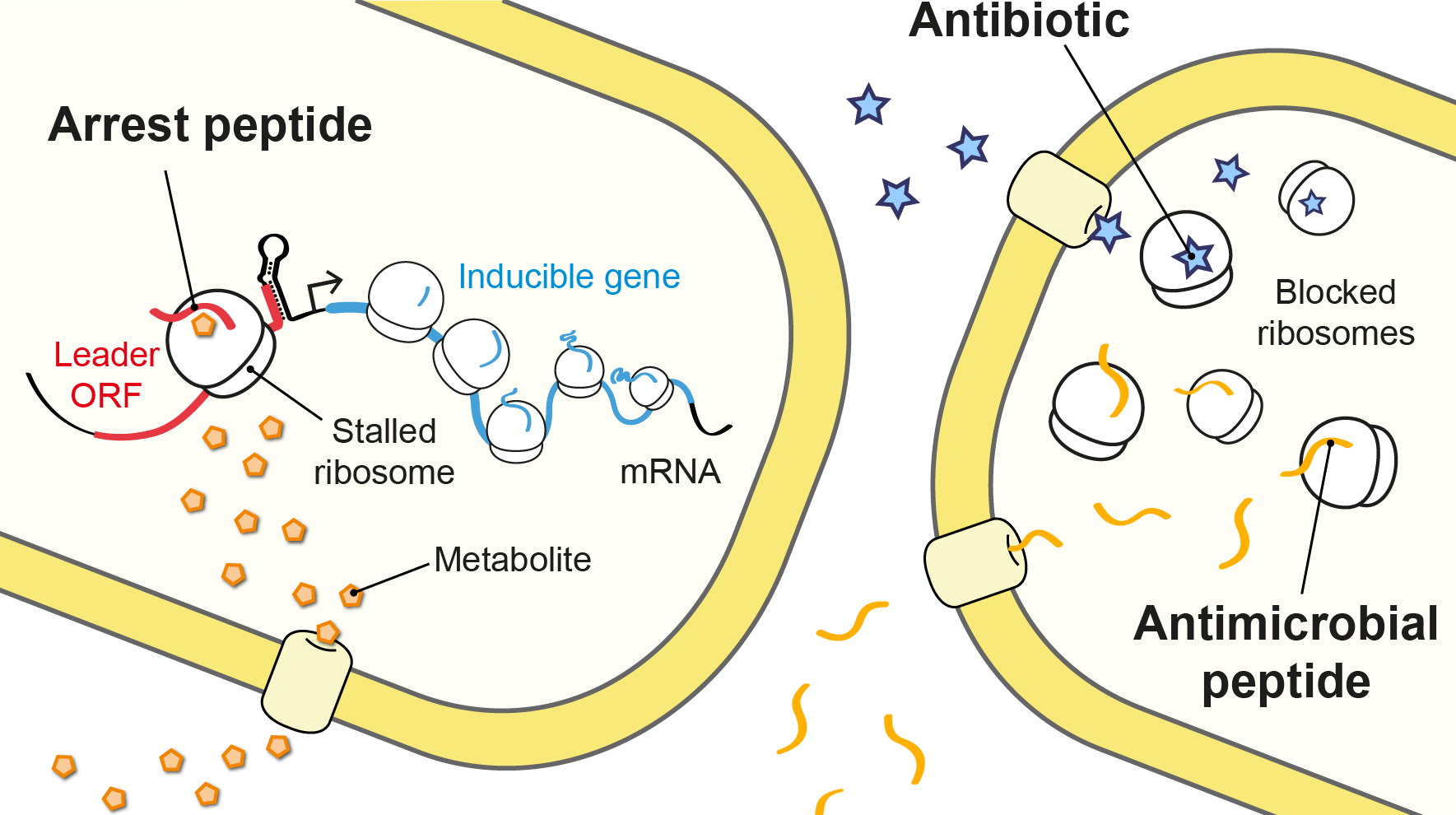
The molecules we study in the lab. Metabolite-sensing arrest peptides, antimicrobial peptides and antibiotics.
Ribosomes are the large macromolecular complexes responsible for accurately translating the genetic information contained in messenger RNA into protein. Our group seeks to understand how ribosomes make proteins, how their activity is regulated in response to different stimuli, and how a variety of small molecules are able to block these complex molecular machines.
Our main focus is the bacterial ribosome and how it is affected by nascent proteins known as arrest peptides, antimicrobial peptides produced by the host immune response, and antibiotics that target the translational machinery.
To understand the molecular mechanisms at play, we use a combination of structural biology (cryo-EM), high-throughput functional characterization and biochemistry.
Research areas
During translation, nascent proteins pass through a long cavity spanning the large subunit of the ribosome – known as the nascent polypeptide exit tunnel – before being released into the cytoplasm or delivered to the protein translocation machinery.
Although most proteins can easily complete this journey, interactions between specific nascent amino acid sequences and the exit tunnel result in impaired translation and ribosome stalling on the messenger RNA. These arrest peptides often block their own translation in response to a small molecule, such as a drug or metabolite, which is sensed by the ribosome nascent chain complex. Thus, arrest peptides are used for metabolite-dependent gene regulation in both prokaryotes and eukaryotes (Seip & Innis, 2016).
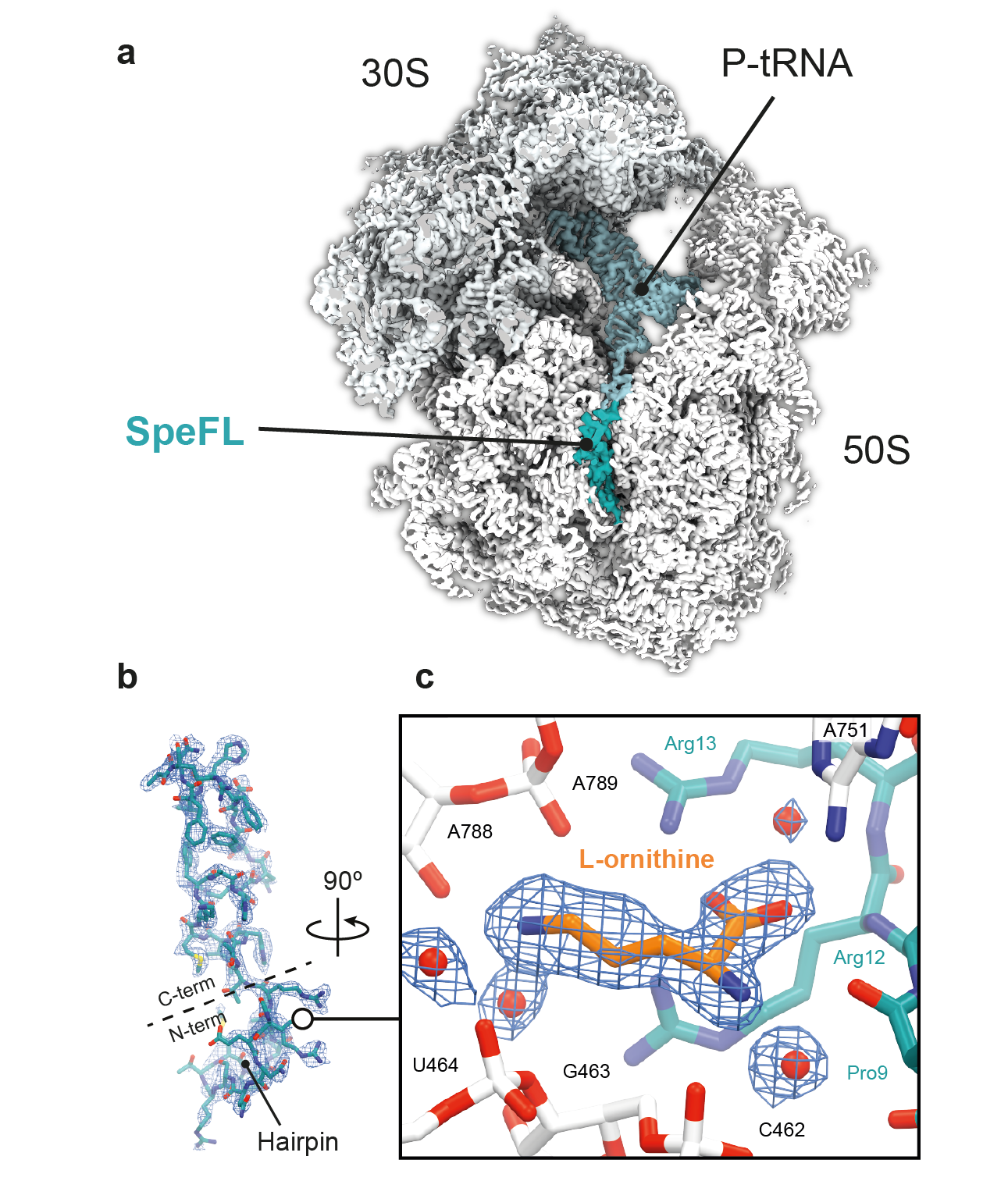
One of our group’s main objectives is the large-scale identification and characterization of arrest peptides in bacteria, which we believe constitute a significant fraction of the hidden proteome of bacteria – the collection of short open reading frames (<60 amino acids) that have escaped genome annotation to date. For example, we have recently identified a new arrest peptide in γ-proteobacteria, called SpeFL, and have shown that the capture of a single molecule of the amino acid ornithine by a ribosome translating SpeFL is sufficient to activate polyamine biosynthesis. Using cryo-EM, we have shown that the bacterial ribosome and SpeFL form a highly selective binding pocket for L-ornithine, capable of discriminating between this amino acid and near-cognate ligands (Herrero del Valle et al., 2020).
Structural basis for the specific recognition of L-ornithine by a SpeFL–70S complex. a, Transverse section of a cryo-EM density map of the SpeFL–70S complex, with the SpeFL peptide and tRNA in turquoise. b, Cryo-EM density and molecular model of SpeFL. c, Binding pocket formed by the ribosome (white) and SpeFL (turquoise), with a single L-ornithine molecule (orange) surrounded by solvent molecules (red) fitted into the cryo-EM density.
This and earlier studies have also shown that arrest peptides block translation by interfering with key aspects of ribosome function, such as tRNA accommodation, peptide bond formation or peptide release. However, the arrest code that determines whether a given nascent peptide is likely to inhibit its own synthesis remains to be elucidated, the range of metabolites that can be detected by the nascent peptide is unknown, and the molecular basis of metabolite sensing by the ribosome is only just beginning to be understood.
Using cryo-EM and high-throughput tools developed in-house – such as inverse toeprinting (Seip et al., 2018) – were are systematically addressing these questions in order to reveal the true extent of metabolite sensing by arrest peptides.
Proline-rich antimicrobial peptides (PrAMPs) produced by the host immune response of insects and mammals display potent antimicrobial activity against Gram-negative bacteria and therefore represent a promising avenue for antibiotic development.
Although PrAMPs such as oncocin (from the milkweed bug) or bactenecin-7 (from cows) were first thought to inhibit bacterial growth by binding to the chaperone protein DnaK, recent studies have shown that they have much greater affinity for the bacterial ribosome. However, their detailed mode of action remained unclear.
In order to determine how these peptides inhibit translation, we determined the crystal structures of five different PrAMPs in complex with the Thermus thermophilus 70S ribosome. Our structures showed that these host defense molecules inhibit the transition from the initiation phase to the elongation phase of translation by binding to the nascent polypeptide exit tunnel and peptidyl transferase center of the ribosome (Seefeldt et al., 2015, Seefeldt et al., 2016, Mardirossian et al., 2018). Since these natural compounds share structural similarities with arrest peptides, the latter may prove helpful in steering the search for new peptide-based antimicrobials that are effective against resistant pathogens.
Using structural biology, droplet-based microfluidics, and biochemistry, we continue to explore the mechanisms by which naturally occurring antimicrobial compounds target and inhibit the bacterial translational machinery.
Ribosome inhibition by antimicrobial peptides. The insect-derived proline-rich antimicrobial peptide Onc112 inhibits bacterial protein synthesis by blocking and destabilizing the translation initiation complex. Other PrAMPs like Bac7, metalnikowin or pyrrhocoricin operate through a similar mechanism.
The threat of bacteria resistant to multiple antibiotics is a major public health challenge that must be tackled through coordinated action on multiple levels. As infectious pathogens have become increasingly resistant to the available drugs, antibiotic discovery programs in major pharmaceutical companies have struggled to produce new antibiotic scaffolds capable of sidestepping current resistance mechanisms. Therefore, new strategies are needed to secure a steady supply of scaffolds and counter the spread of resistance.
The bacterial ribosome is a target for several major classes of antibiotics, including molecules that block peptide bond formation (chloramphenicol, oxazolidinones), impede the synthesis and movement of nascent proteins through the exit tunnel (macrolides), interfere with the decoding of messenger RNA (aminoglycosides) or prevent the translocation of tRNAs during the elongation step of protein synthesis (tuberactinomycins).
Using structural biology and high-throughput approaches, we are (i) characterizing bacterial arrest peptides responsible for the induction of erm resistance genes in response to macrolide antibiotics, and (ii) revisiting the mechanisms of action of ribosome-targeting antibiotics, focusing on the defined functional states they target. This includes re-examining poorly characterized natural products from the golden age of antibiotic discovery (1950-1960s) to identify the detailed molecular mechanisms by which they block translation. A better understanding of how these antibiotics work could help design improved molecules that are effective against resistant pathogens.
In addition, we are developing high-throughput approaches that use the bacterial ribosome as a platform for the production and selection of translation inhibitors (Charon et al., 2019). We are particularly interested in molecules that target new sites on the ribosome, as they could be used as scaffolds to design novel drugs capable of bypassing known resistance mechanisms.
In order to understand how peptides or antibiotics inhibit peptide bond formation, we must first have a clear picture of the mechanism by which ribosomes catalyze peptidyl transfer.
Peptide bond formation takes place within an active site that is composed primarily of RNA. Our high-resolution structures of the bacterial ribosome in complex with full-length tRNA substrates reveal a network of hydrogen bonds (or “proton wire”) along which proton transfer could take place to assist catalysis (Polikanov et al. (2014)). This has led us to propose a mechanism for peptide formation in which the ribosome together with the A- and P-tRNAs trigger the reaction by activating a water molecule. As this proposed catalytic water is cut off from the bulk solvent by the N-terminus of ribosomal protein L27 in bacteria, we are currently investigating a possible regulatory role for this protein during translation.
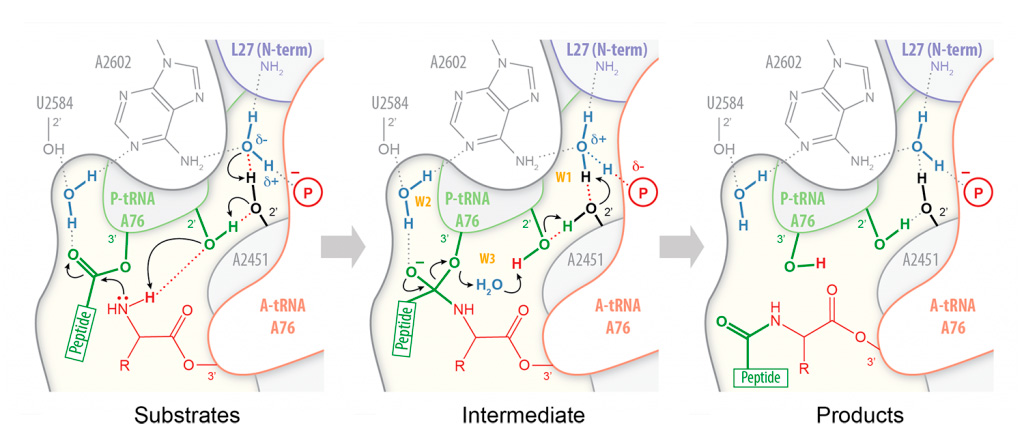
Proton wire model for peptide bond formation. In our proposed mechanism for peptide bond formation, nucleophilic attack is facilitated through the deprotonation of the a-amine of the incoming amino acid by a catalytic water molecule (W1) positioned at the extremity of a “proton wire”.


